-Search query
-Search result
Showing 1 - 50 of 172 items for (author: lewis & k)

PDB-8qox: 
Two-component assembly of SlaA and SlaB S-layer proteins of Sulfolobus acidocaldarius
Method: subtomogram averaging / : Gambelli L, McLaren M, Isupov M, Conners R, Daum B

PDB-8qp0: 
A hexamer pore in the S-layer of Sulfolobus acidocaldarius formed by SlaA protein
Method: subtomogram averaging / : Gambelli L, McLaren M, Isupov M, Conners R, Daum B

EMDB-18127: 
S-layer of archaeon Sulfolobus acidocaldarius by subtomogram averaging
Method: subtomogram averaging / : Gambelli L, McLaren MJ, Daum B

EMDB-15530: 
S-layer protein SlaA from Sulfolobus acidocaldarius at pH 10.0
Method: single particle / : Gambelli L, Isupov MN, Daum B

EMDB-15531: 
S-layer protein SlaA from Sulfolobus acidocaldarius at pH 7.0
Method: single particle / : Gambelli L, Isupov MN, Daum B

PDB-8an2: 
S-layer protein SlaA from Sulfolobus acidocaldarius at pH 10.0
Method: single particle / : Gambelli L, Isupov MN, Daum B

PDB-8an3: 
S-layer protein SlaA from Sulfolobus acidocaldarius at pH 7.0
Method: single particle / : Gambelli L, Isupov MN, Daum B

EMDB-27703: 
Structure of RBD directed antibody DH1047 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interace
Method: single particle / : May AJ, Manne K, Acharya P

PDB-8dtk: 
Structure of RBD directed antibody DH1047 in complex with SARS-CoV-2 spike: Local refinement of RBD-Fab interace
Method: single particle / : May AJ, Manne K, Acharya P

EMDB-41182: 
Cryo-EM map of the Unmodified nucleosome core particle in 100 mM KCl with local resolution values
Method: single particle / : Huang SK, Kay LE, Rubinstein JL
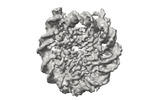
EMDB-41183: 
Cryo-EM map of the PARylated nucleosome core particle in 100 mM KCl with local resolution values
Method: single particle / : Huang SK, Kay LE, Rubinstein JL
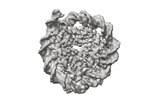
EMDB-41184: 
Cryo-EM map of the Unmodified nucleosome core particle in 5 mM KCl with local resolution values
Method: single particle / : Huang SK, Kay LE, Rubinstein JL
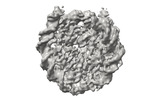
EMDB-41178: 
Cryo-EM map of the PARylated nucleosome core particle in 5 mM KCl with local resolution values
Method: single particle / : Huang SK, Kay LE, Rubinstein JL

EMDB-16519: 
Slipper limpet hemocyanin didecamer
Method: single particle / : Pasqualetto G, Young MT

EMDB-16523: 
Slipper limpet hemocyanin tridecamer
Method: single particle / : Clare D, Young MT

EMDB-14635: 
S-layer protein SlaA from Sulfolobus acidocaldarius at pH 4.0
Method: single particle / : Gambelli L, Isupov MN, Daum B

PDB-7zcx: 
S-layer protein SlaA from Sulfolobus acidocaldarius at pH 4.0
Method: single particle / : Gambelli L, Isupov MN, Daum B
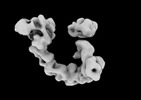
EMDB-29101: 
EGFR:Degrader:VHL:Elongin-B/C:Cul2
Method: single particle / : Rosenberg SC

EMDB-28754: 
Client-bound structure of a DegP trimer within a 12mer cage
Method: single particle / : Harkness RW, Ripstein ZA, Di Trani JM, Kay LE

EMDB-28781: 
Structure of a 12mer DegP cage bound to the client protein hTRF1
Method: single particle / : Harkness RW, Ripstein ZA, Di Trani JM, Kay LE

EMDB-28800: 
Structure of an 18mer DegP cage bound to the client protein hTRF1
Method: single particle / : Harkness RW, Ripstein ZA, Di Trani JM, Kay LE

EMDB-28801: 
Structure of a 24mer DegP cage bound to the client protein hTRF1
Method: single particle / : Harkness RW, Ripstein ZA, Di Trani JM, Kay LE

EMDB-28806: 
Structure of a 30mer DegP cage bound to the client protein hTRF1
Method: single particle / : Harkness RW, Ripstein ZA, Di Trani JM, Kay LE

EMDB-28808: 
Structure of a 60mer DegP cage bound to the client protein hTRF1
Method: single particle / : Harkness RW, Ripstein ZA, Di Trani JM, Kay LE

PDB-8f0a: 
Client-bound structure of a DegP trimer within a 12mer cage
Method: single particle / : Harkness RW, Ripstein ZA, Di Trani JM, Kay LE

PDB-8f0u: 
Structure of a 12mer DegP cage bound to the client protein hTRF1
Method: single particle / : Harkness RW, Ripstein ZA, Di Trani JM, Kay LE

PDB-8f1t: 
Structure of an 18mer DegP cage bound to the client protein hTRF1
Method: single particle / : Harkness RW, Ripstein ZA, Di Trani JM, Kay LE

PDB-8f1u: 
Structure of a 24mer DegP cage bound to the client protein hTRF1
Method: single particle / : Harkness RW, Ripstein ZA, Di Trani JM, Kay LE

PDB-8f21: 
Structure of a 30mer DegP cage bound to the client protein hTRF1
Method: single particle / : Harkness RW, Ripstein ZA, Di Trani JM, Kay LE

PDB-8f26: 
Structure of a 60mer DegP cage bound to the client protein hTRF1
Method: single particle / : Harkness RW, Ripstein ZA, Di Trani JM, Kay LE

EMDB-25994: 
Structure of the rabbit 80S ribosome stalled on a 2-TMD Rhodopsin intermediate in complex with the multipass translocon
Method: single particle / : Kim MK, Lewis AJO, Keenan RJ, Hegde RS

EMDB-26133: 
Structure of the rabbit 80S ribosome stalled on a 4-TMD Rhodopsin intermediate in complex with the multipass translocon
Method: single particle / : Kim MK, Lewis AJO, Keenan RJ, Hegde RS

PDB-7tm3: 
Structure of the rabbit 80S ribosome stalled on a 2-TMD Rhodopsin intermediate in complex with the multipass translocon
Method: single particle / : Kim MK, Lewis AJO, Keenan RJ, Hegde RS

PDB-7tut: 
Structure of the rabbit 80S ribosome stalled on a 4-TMD Rhodopsin intermediate in complex with the multipass translocon
Method: single particle / : Kim MK, Lewis AJO, Keenan RJ, Hegde RS
PDB-7t3h: 
MicroED structure of Dynobactin
Method: electron crystallography / : Yoo BK, Kaiser JT, Rees DC, Miller RD, Iinishi A, Lewis K, Bowman S

EMDB-14242: 
E. coli BAM complex (BamABCDE) bound to dynobactin A
Method: single particle / : Jakob RP, Hiller S

PDB-7r1w: 
E. coli BAM complex (BamABCDE) bound to dynobactin A
Method: single particle / : Jakob RP, Hiller S, Maier T

EMDB-13978: 
S. cerevisiae CMGE nucleating origin DNA melting
Method: single particle / : Lewis JS, Sousa JS, Costa A

EMDB-13988: 
S. cerevisiae CMGE dimer nucleating origin DNA melting
Method: single particle / : Lewis JS, Sousa JS, Costa A

EMDB-14439: 
S. cerevisiae CMGE dimer nucleating origin DNA melting
Method: single particle / : Lewis JS, Sousa JS, Costa A

PDB-7qhs: 
S. cerevisiae CMGE nucleating origin DNA melting
Method: single particle / : Lewis JS, Sousa JS, Costa A

PDB-7z13: 
S. cerevisiae CMGE dimer nucleating origin DNA melting
Method: single particle / : Lewis JS, Sousa JS, Costa A

EMDB-25408: 
Mfd DNA complex
Method: single particle / : Oakley AJ, Xu ZQ

EMDB-11953: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Composite Map
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-11954: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 2)
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-14810: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Consensus Map
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-14811: 
SARS-CoV-2 S 2P trimer in complex with monovalent DARPin R2 (State 1) - Focused Refinement
Method: single particle / : Hurdiss DL, Drulyte I

EMDB-12676: 
Vibrio vulnificus stressosome
Method: single particle / : Kaltwasser S, Heinz V

EMDB-11971: 
Stressosome complex from Listeria innocua
Method: single particle / : Miksys A, Fu L, Madej MG, Ziegler C
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search




 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
